The prokaryotic type II CRISPR/Cas9 system has revolutionized the field of genome engineering, allowing researchers to edit the genomes of a great variety of organisms with unprecedented speed and precision. Experimental approaches based on this highly precise and versatile system have the potential to accelerate the functional identification and detailed characterization of cancer genes. We have recently demonstrated the feasibility of utilizing this powerful approach to model liver cancer by directly mutating cancer genes in vivo (Xue et al., 2014). Furthermore, we have recently adapted this technology to model lung cancer mutations in vivo utilizing pre-clinical genetically engineered mouse models (GEMMs) of lung adenocarcinoma (Sánchez-Rivera et al., 2014). We are currently extending these CRISPR-based in vivo approaches to other GEMMs of human cancer, such as small cell lung cancer, squamous cell carcinoma and pancreatic cancer, among others. In addition, we are extending the utility of these approaches to model both focal and chromosomal alterations frequently observed in human cancer. As the field of cancer genomics moves forward with the identification and cataloguing of recurrent somatic mutations and copy number alterations in a variety of human cancers, our CRISPR-based experimental platforms offer a rapid, precise and affordable opportunity to functionally interrogate the cancer genome using sophisticated GEMMs of human cancer.
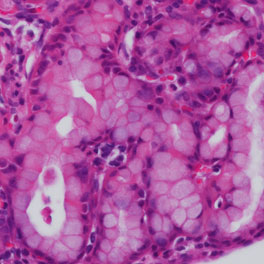
CRISPR-mediated disruption of Nkx2-1 in vivo leads to mucinous adenocarcinoma, typified by the presence of mucin-producing cells.

